Machine Learning
The goal of BioSense’s AI Group is to get a comprehensive and accurate overview of biosystems, make phenotyping and plant breeding more efficient, understand molecular biology better and offer optimal and automatic decision-making to the farmer. Our research starts with the problem and requires innovative application of state-of-the-art algorithms and development of the novel ones. Machine learning algorithms extensively used in BioSense applications include Random Forest, SVM, Decision Trees, Ridge Regression, Spectral Clustering and many others including special family of machine learning algorithms- Deep Learning, and where the complexity of the problem goes beyond the available algorithms, BioSense researchers develop the novel ones.
Example of novel algorithm is include weighted histograms regression (WHR). WHR is based on a voting mechanism, where training samples are voting for the output value of a test sample. The votes are weighted according to the similarity of features and used to form a histogram that essentially represents the PDF of the output, while the expectation of the PDF is essentially the predicted value. This algorithm proved very useful for yield prediction, where it outperformed random forest, SVM and artificial neural networks.
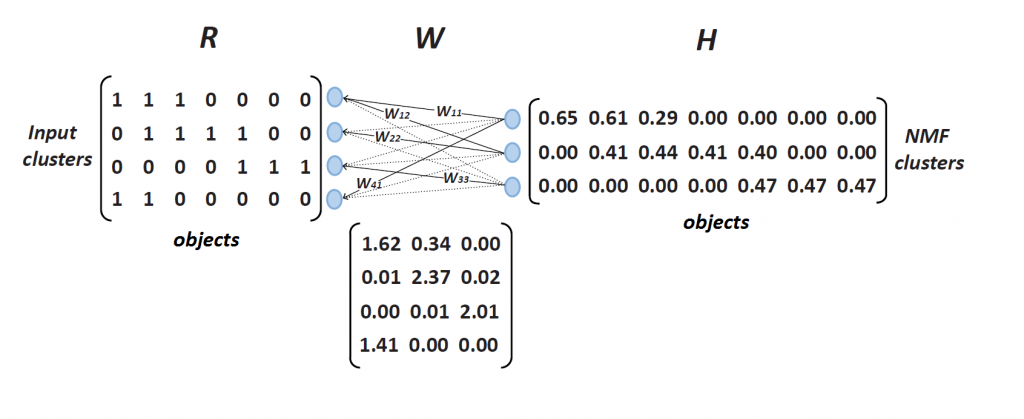
Another novel algorithm is integrative clustering based on Nonnegative matrix factorization. Combining results of individual clusterings together into an ensemble leverages evidence accumulation in order to improve the results of clustering. Individual clustering results are merged into a joint cluster membership matrix that is further factorized into matrices of encodings and basis. Algorithm can integrate clusterings stemming from different data sets, different data preprocessing steps or different subsamples of features or objects.
This algorithm proved very useful for functional genomics. Proposed approach based on nonnegative matrix factorization can fuse diverse data sources and infer gene clusters with high functional enrichment and improved gene coverage.